

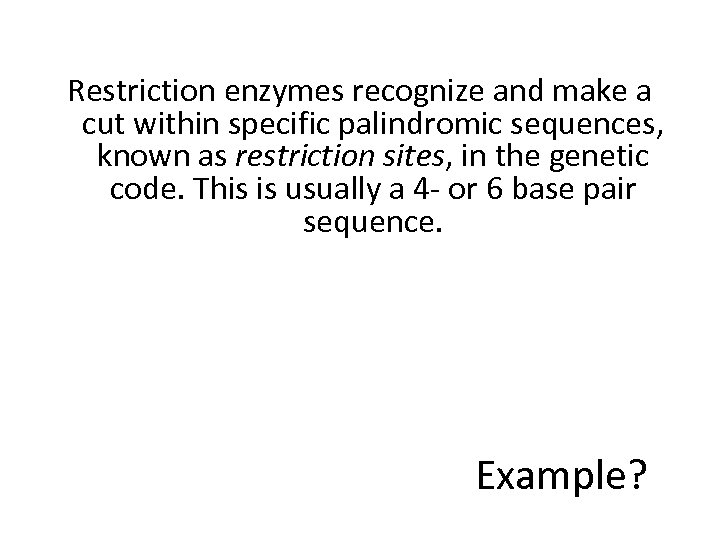
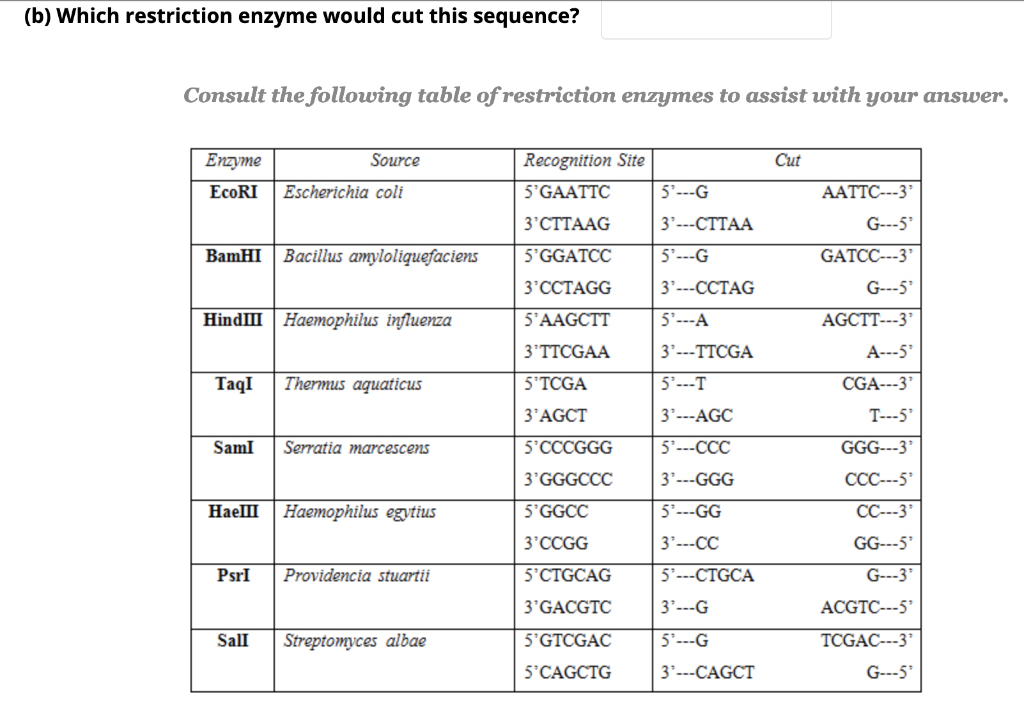
Not sure why Larry Parnell was down-voted, he was not technically wrong.Ĭrystal structures of the most popular restriction enzymes are already known and can easily be found the Protein Data Bank or Wikipedia for graphical reference. If you look at the histogram of lengths, there is no bias towards even recognition site lengths except for length 6. Ggplot(df, aes(x=mids, y=counts, fill=even))+geom_bar(stat="identity")
P=0.05, 1 Degree of Freedom: Critical Value of 3.841
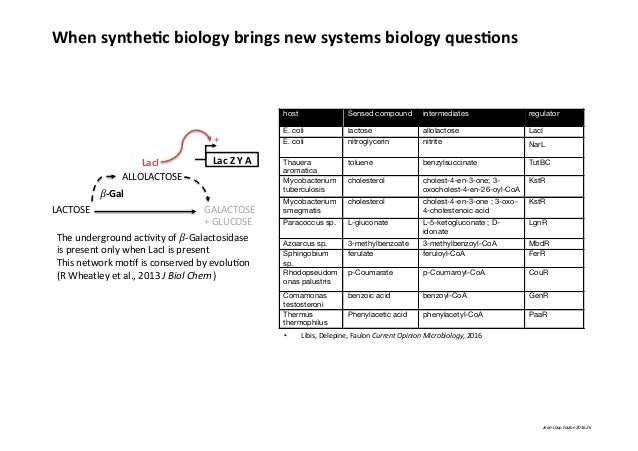
There is no significant difference between the number of restriction sequences The results are probably best summarised graphically: I wondered if this was just a co-incidence, so I took the data from this site for over a thousand known recognition sites and put it into a spreadsheet ( XLS uploaded here). When reading my textbook I noticed that in all examples but one from eight the recognition site was an even number of bases.


 0 kommentar(er)
0 kommentar(er)
